In the pursuit to determine the optimum length of chopsticks, two laboratory studies were conducted, using a randomised complete block design, to evaluate the effects of the length of the chopsticks on the food-serving performance of adults and children. Thirty-one male junior college students and 21 primary school pupils served as subjects for the experiment.The response was recorded in terms of food-pinching efficiency (number of peanuts picked, and placed in a cup).
Data source https://www.udacity.com/api/nodes/4576183932/supplemental_media/chopstick-effectivenesscsv/download
The data was read in using read.csv() and allotted appropriate column names.
stick<-read.csv(file.choose(),header = TRUE,stringsAsFactors = FALSE)
chop<-stick
names(chop)<-c("Efficiency","Individual","Length")
Next, we’ll convert the ‘Length’ and ‘Individual’ to factors and the dependent variable ‘Efficiency’ to numeric to carry out our repeated measures ANOVA analysis.
chop$Efficiency<-as.numeric(chop$Efficiency) chop$Length<- factor(chop$Length) chop$Individual<-factor(chop$Individual)
Let’s use the tapply() to find out the mean pinching efficiency grouped by the chopstick length and plot the result.
group_mean<-tapply(chop$Efficiency,chop$Length,mean) plot(group_mean,type="p",xlab="Chopstick Length",ylab="Mean Efficiency", main="Average Food Pinching Efficiency \n by Chopstick Length",col="green",pch=16)
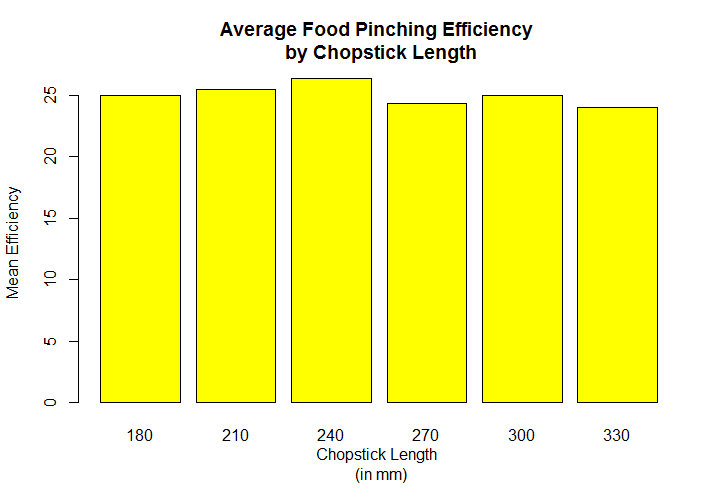
Visually, we can see that the efficiency grows steadily from 180 mm through to the 240 and then falling sharply at 270 mm and despite a slight increase at 300,it continues to fall at 330.
Before we can perform the repeated measures ANOVA, we will need to check for the Sphericity which assumes the homogeneity of variance among differences between all possible pairs of groups. This is done by the Mauchly’s test.
chop1<- cbind(chop$Efficiency[chop$Length==180],
chop$Efficiency[chop$Length==210],
chop$Efficiency[chop$Length==240],
chop$Efficiency[chop$Length==270],
chop$Efficiency[chop$Length==300],
chop$Efficiency[chop$Length==330])
mauchly.test (lm (chop1 ~ 1), X = ~1)
> mauchly.test (lm (chop1 ~ 1), X = ~1)
Mauchly's test of sphericity
Contrasts orthogonal to
~1
data: SSD matrix from lm(formula = chop1 ~ 1)
W = 0.43975, p-value = 0.05969
Since, (thankfully!) the p-value is >0.05, the homogeneity of variance assumption holds and no further adjustment is necessary.
We can now get on with the repeated measures ANOVA.
aov.chop = aov(Efficiency~Length + Error(Individual/Length),data=chop)
summary(aov.chop)
Error: Individual
Df Sum Sq Mean Sq F value Pr(>F)
Residuals 30 2278 75.92
Error: Individual:Length
Df Sum Sq Mean Sq F value Pr(>F)
Length 5 106.9 21.372 5.051 0.000262 ***
Residuals 150 634.6 4.231
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
The F-ratio is 5.051 and p-value is 0.000262 which means there is significant effect of the length of the chopstick on the eating efficiency.
Since, the F-ratio is significant , we can now process the post-hoc analysis to perform pair-wise comparisons.
1. Holm’s adjustment
with(chop,pairwise.t.test(Efficiency,Length,paired=T))
> with(chop,pairwise.t.test(Efficiency,Length,paired=T))
Pairwise comparisons using paired t tests
data: Efficiency and Length
180 210 240 270 300
210 1.000 - - - -
240 0.323 1.000 - - -
270 1.000 0.034 0.035 - -
300 1.000 1.000 0.198 1.000 -
330 0.630 0.048 8.1e-05 1.000 0.435
P value adjustment method: holm
2. Bonferroni adjustment
with(chop,pairwise.t.test(Efficiency,Length,paired=T,p.adjust.method="bonferroni"))
> with(chop,pairwise.t.test(Efficiency,Length,paired=T,p.adjust.method="bonferroni"))
Pairwise comparisons using paired t tests
data: Efficiency and Length
180 210 240 270 300
210 1.000 - - - -
240 0.485 1.000 - - -
270 1.000 0.036 0.040 - -
300 1.000 1.000 0.269 1.000 -
330 1.000 0.060 8.1e-05 1.000 0.725
P value adjustment method: bonferroni
As we can see, the Bonferroni adjustment inflates the p-values as compared to Holm’s adjustment.
3. Treatment-by-Subjects
Another method which can be employed is the Treatment-by-Subjects i.e. as a 2 factor design without interaction , without replication. The advantage of this approach is that it allows us to employ the TukeyHSD() post-hoc .
aov.chop1 = aov(Efficiency ~ Length + Individual, data=chop)
summary(aov.chop1)
> summary(aov.chop1)
Df Sum Sq Mean Sq F value Pr(>F)
Length 5 106.9 21.37 5.051 0.000262 ***
Individual 30 2277.5 75.92 17.944 < 2e-16 ***
Residuals 150 634.6 4.23
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
>
TukeyHSD(aov.chop1,which="Length")
> tukey
Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = Efficiency ~ Length + Individual, data = chop)
$Length
diff lwr upr p adj
210-180 0.54870968 -0.9595748 2.05699418 0.8999148
240-180 1.38774194 -0.1205426 2.89602644 0.0904885
270-180 -0.61129032 -2.1195748 0.89699418 0.8503866
300-180 0.03290323 -1.4753813 1.54118773 0.9999999
330-180 -0.93548387 -2.4437684 0.57280063 0.4749602
240-210 0.83903226 -0.6692522 2.34731676 0.5959492
270-210 -1.16000000 -2.6682845 0.34828450 0.2346843
300-210 -0.51580645 -2.0240910 0.99247805 0.9213891
330-210 -1.48419355 -2.9924781 0.02409096 0.0565555
270-240 -1.99903226 -3.5073168 -0.49074775 0.0025803
300-240 -1.35483871 -2.8631232 0.15344579 0.1053005
330-240 -2.32322581 -3.8315103 -0.81494130 0.0002412
300-270 0.64419355 -0.8640910 2.15247805 0.8199855
330-270 -0.32419355 -1.8324781 1.18409096 0.9893780
330-300 -0.96838710 -2.4766716 0.53989741 0.4349561
By studying the difference between the means and the Tukey adjusted p-value, the most significant diff is between the groups 240-330 , with a mean difference of 2.32.
Conclusion:
The results show that the food-pinching performance is significantly affected by the length of the chopsticks, and that chopsticks of about 240 mm long performed the best.